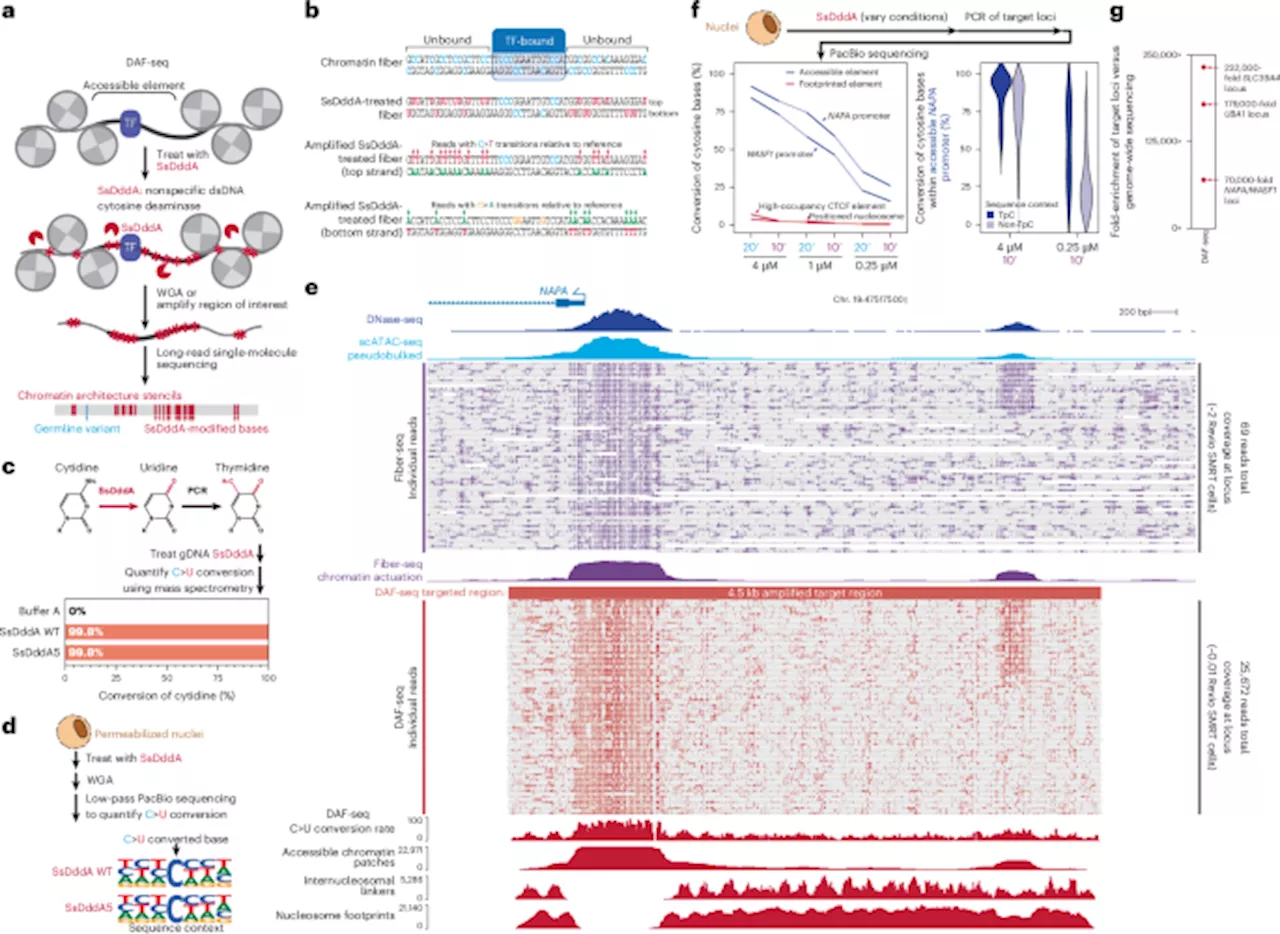
Researchers at the University of Washington have developed a groundbreaking technology called Deaminase-Assisted single-molecule chromatin Fiber sequencing (DAF-seq). This innovative approach allows for the mapping of chromatin fiber architectures at a near-nucleotide resolution, unveiling the complexities of gene regulation in diploid organisms. The study addresses a significant gap in understanding the variability in protein occupancy across different haplotypes and individual cells.
DAF-seq provides unprecedented insights into cooperative protein interactions along chromosome-length chromatin fibers. By generating maps of protein co-occupancy across approximately 99% of each cell’s mappable genome, the technology reveals intricate patterns of regulatory element activation. This level of detail is achieved while also profiling DNA sequences, allowing researchers to assess the functional impact of somatic variants and rare chromatin epialleles.
Revolutionizing Gene Regulation Studies
The findings demonstrate that chromatin plasticity varies significantly within and between diploid cells. For instance, the study found that chromatin actuation diverges by 61% between haplotypes within a single cell and 63% between different cells. These variations highlight the dynamic nature of chromatin and its role in gene expression regulation.
Additionally, the research indicates that regulatory elements tend to activate cooperatively along the same chromatin fiber. This activation appears to follow a distance-dependent pattern that resembles the interactions mediated by cohesin loops, suggesting a complex network of regulatory mechanisms operating at the chromatin level.
Lead author A.B. Stergachis emphasized that DAF-seq not only enhances the understanding of protein occupancy across entire chromosomes but also does so with single-nucleotide, single-molecule, single-haplotype, and single-cell precision. This level of granularity can significantly advance the field of gene regulation and epigenetics.
Implications for Future Research
The implications of DAF-seq extend beyond fundamental research. The technology has the potential to inform therapeutic strategies for various diseases by elucidating how genetic variations influence chromatin behavior and gene expression. The ability to understand these mechanisms could lead to targeted interventions in conditions where gene regulation is disrupted.
This research received support from the National Institutes of Health Common Fund and the Chan Zuckerberg Initiative, among others. A.B. Stergachis holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is recognized as a Pew Biomedical Scholar, underscoring the credibility of the research team.
As the scientific community continues to explore the complexities of gene regulation, DAF-seq stands out as a pivotal tool. Its ability to provide detailed insights into the chromatin landscape could pave the way for new discoveries in genetics and personalized medicine.