URGENT UPDATE: A groundbreaking deep learning model has been unveiled, revolutionizing the analysis of fruit tissue microstructure. Published on July 5, 2025, by a team from KU Leuven led by Pieter Verboven, this innovative approach significantly outperforms traditional 2D methods in accuracy and efficiency.
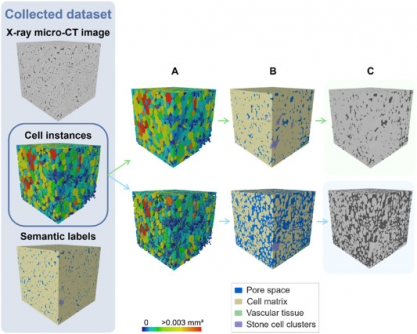
The new model, designed for analyzing apple and pear tissues, utilizes advanced X-ray micro-CT technology to achieve precise 3D segmentation of plant tissues. Traditional microscopy methods require extensive preparation and are limited in field size, but this model allows for non-destructive imaging, making it a game-changer for agricultural research and food science.
This 3D deep learning framework not only automates the labeling and quantification of plant tissue architecture but also enhances our understanding of vital metabolic processes. The model’s impressive performance reached an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, surpassing previous 2D approaches. It achieves nearly flawless segmentation of pore spaces and cell matrices, with a Dice Similarity Coefficient (DSC) of up to 0.90 for stone cell clusters.
“This model provides plant scientists with a powerful, non-destructive tool for studying how microscopic structures influence water, gas, and nutrient transport,”
said Verboven. The implications of this research stretch far beyond mere academic interest; it promises to accelerate the pace of discoveries in fruit storage behavior and physiological disorders such as browning or watercore.
With its ability to integrate seamlessly into existing X-ray micro-CT instruments, this model makes advanced artificial intelligence accessible for plant anatomy research. It empowers scientists to delve deeper into the cellular arrangements that dictate fruit texture, storability, and overall quality.
The study leveraged a 3D panoptic segmentation framework built on the 3D extension of Cellpose and a 3D Residual U-Net. This sophisticated setup was trained on augmented datasets, enhancing its ability to classify various tissue types. Despite challenges with dataset imbalance, the model’s outcomes demonstrate a significant leap forward in quantifying fruit tissue microstructure.
As the agricultural and scientific communities eagerly await further applications of this technology, the potential for improving crop management and yield optimization is immense. Researchers can now explore diverse crops, studying tissue development and responses to environmental stressors more effectively than ever before.
The significance of this research cannot be overstated. It paves the way for a new era in plant research, where deep learning and AI combine to unlock the secrets of plant physiology. The ability to produce accurate, automated tissue analysis not only reduces manual labor but also enhances the precision of tissue characterization.
As the study gains traction, experts anticipate that the findings will catalyze further innovations in plant phenomics, impacting agriculture and horticulture globally. The integration of this deep learning model into everyday research practices promises to accelerate discoveries that can improve food security and sustainability.
Stay tuned for more updates as this exciting research unfolds, and anticipate a future where AI drastically transforms how we understand and utilize plant biology.